log2(8)[1] 3sin(pi)[1] 1.224647e-162^10[1] 1024sqrt(exp(1))[1] 1.648721What you should know after today:
So, why would one like to learn R?
My personal answer is: it doesn’t matter much whether you learn R, Python or another programming language you can use for statistics and data analysis. The important thing is to learn a programming language, as opposed to using Excel, SPSS or some other point-and-click software.
The most important reason for this is that with a programming language, you solve the problems programmatically, that is by writing some code. This code stays with you, can be inspected, modified, adapted to other needs. That way, you ensure scientific reproducibility, transparency and reusability of your work. Your program, script or Rmarkdown file becomes your lab book, meticulously detailing the steps you have taken to arrive at a given solution. And just like a good lab book can be used to guide you in future experiments, replicating your previous results, you can re-use your code.
Consider the following example: you need to create a complex illustration of your data for a paper manuscript you are working on. You could create the illustration in Excel, modify it in PowerPoint, paste it together with other illustrations and a caption in Word. That is how many people are actually working. However, this approach has several huge disadvantages.
Firstly, if a few months later a reviewer asks you how precisely did you generate the figure, you will have a hard time remembering all the steps that were necessary, all the options you had to click in Excel and so on. Secondly, imagine you receive additional data – you will have to repeat every. Single. Step again. Then of course the figure is not the only one in your paper. What if the editor decides that your color scheme doesn’t fit the journal requirements? You need to go through every single figure and modify the colors. And finally, when you want to create a similar figure for another paper, you will have to start from scratch.
With R or another programming language figuring out how to achieve the same effect might be harder. However, once you have done it, all your operations have been recorded. You can show it to anyone and it will precisely document what you did. You can re-run the generation of your figure automatically. So if your data changes, you simply re-run the code and have your new figure reflect the change. If you want to change the color scheme in your whole paper, it will often be a matter of changing a single line in your code. And finally, for your next paper, you will simply cut, paste and adapt the code you have.
OK, so far so good, but why R in particular, and not, say, Python?
R is not the only language that you can use for data analysis. There are many other languages that are used for this purpose, including Python, Matlab and many others. Each of these languages has its own strengths and weaknesses, and the choice of language depends on your needs. In fact, most bioinformaticians know more than one language, and use the one that is best suited for the task at hand.
I think that R is a particularly good choice for all those who just need a tool to use from time to time to help them with their work. It is relatively easy to learn, it is very powerful, and it has a huge, vibrant, helpful community. However, two other choices are also worth mentioning.
Matlab is a language that is in many ways similar to R. The main differnce is maybe that unlike R, Matlab is not free – it is closed source and you have to pay for a license. This has some advantages. For example, and as you will see during this course, R development is not centralized and so there are many packages that do the same thing. Matlab is in some aspects more consistent and more polished than R, and in some comparisons appears to be faster – and for this, it is often the language of choice for areas such as image analysis.
Python is completely different story. This is a powerful, fast, general purpose programming language. It is more versatile than R, has a much more standardized syntax and development process. However, it is harder to learn and it is not really meant to be used interactively (although it can be – especially when combined with Quarto or Jupyter Notebook). While many statistical modules exist for Python, it is not as strong in this area as R.
R is the name of both, the programming language and of the language interpreter. When you start RStudio, you can see the R language interpreter working in the part of the window left and bottom - called “console”. So yes, you don’t need RStudio to work with R and, in fact, many people prefer to work with R in a different environment.
RStudio is a so called IDE, an Integrated Development Environment. That is, it provides a lot of goodies that help make your work easier, faster and more efficient.
When starting work with a new project, do the following: (i) create a new directory for the project, (ii) open an R script file and save it in the directory you created and (iii) copy necessary data files.
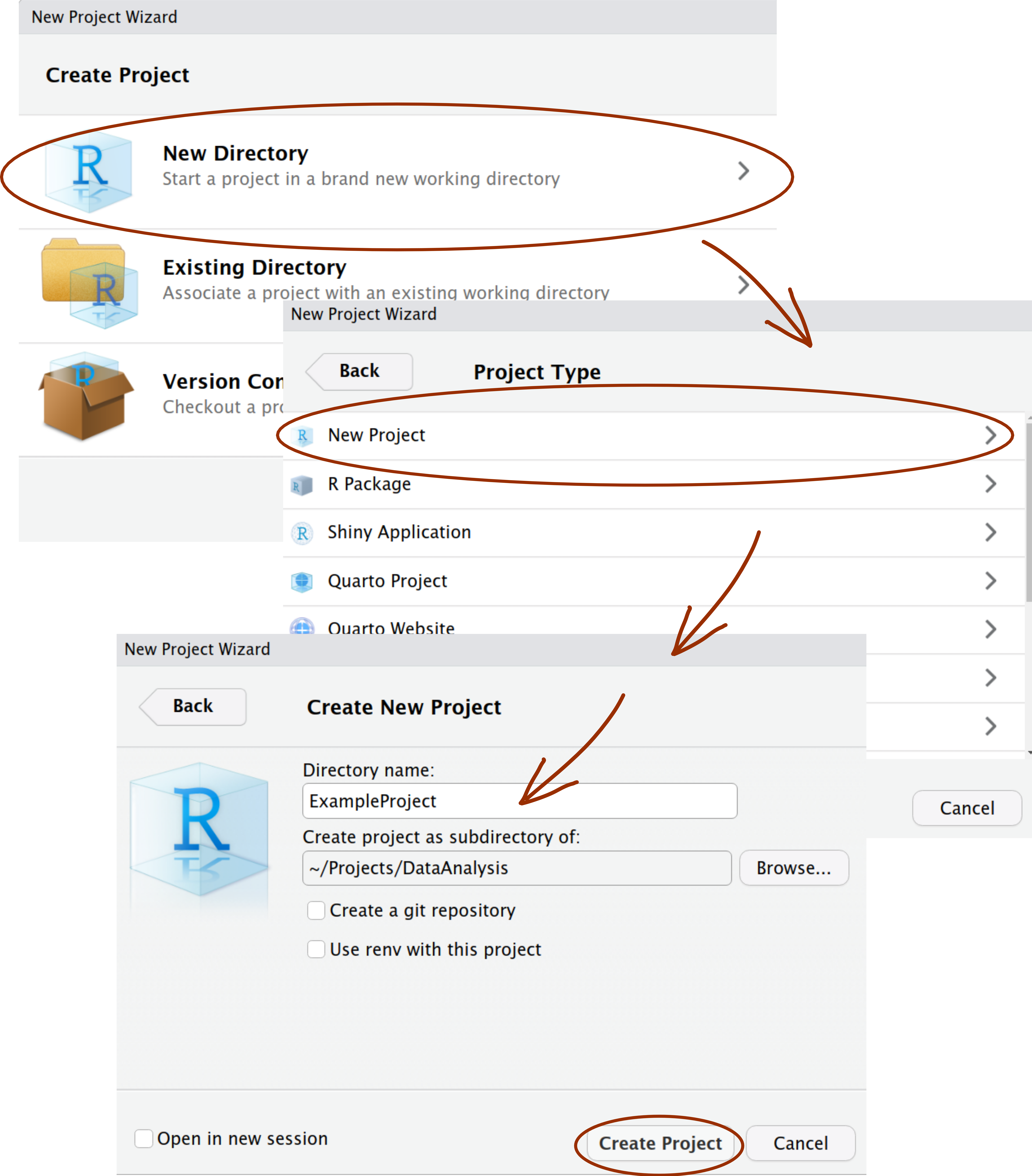
To create a new directory in RStudio, go to File -> New Project. When the dialog window appears, select first “New Directory” and then “New Project”. Click on “Browse…” to select the location where you would like to have the directory created. Enter a name for your project and click on “Create Project”. Presto!
Exercise 1.1
When Rstudio creates a new project, it creates a new directory with the same name as the project. Furthermore, it creates a new file in this directory called projectname.Rproj. This file is used by RStudio to keep track of project-specific settings.
projectname.RprojYou can open this file by double-clicking on it in the Files pane in RStudio. Like most of the files that you will be working with, it is a simple text file: you can open it in any text editor, including RStudio.
The other file I told you to create is a script file. This is where you will later be typing your code, and we will discuss it in more detail in a moment.
Later on, if you choose to do so, R can create two hidden files, Rhistory (called .Rhistory on Unix-like systems and _Rhistory on Windows) and .RData (or _RData). This files save the state of your R session (of your R workspace, to be specific).
.Rhistory and .RDataAs I mentioned before, RStudio is a so-called “integrated development environment” (IDE). It makes working with R much easier and more efficient. You will be exploring many of its components when you start working with R and RStudio, but let us just mention a couple general things. Below is a screenshot of an R session opened in RStudio:
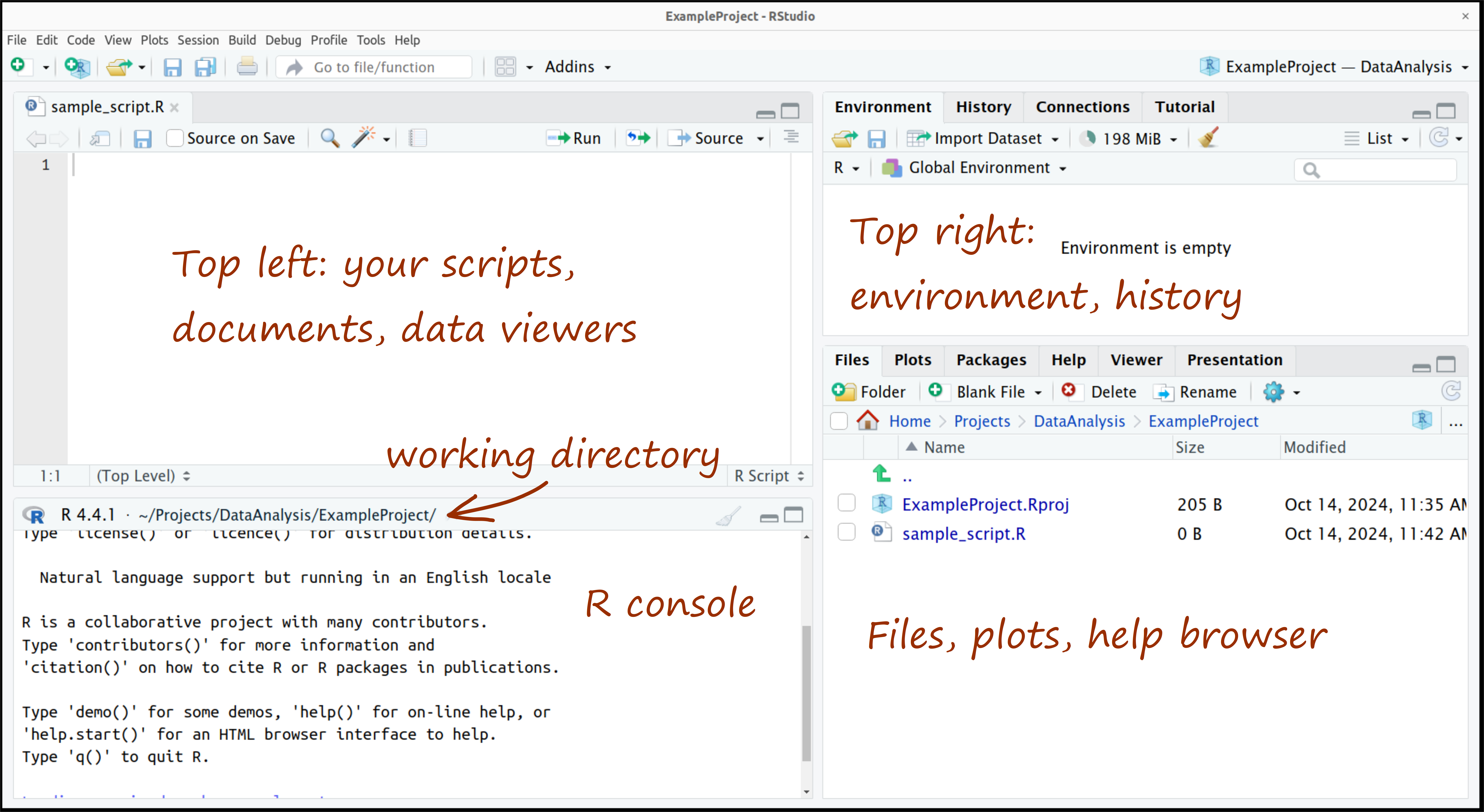
There are four main RStudio panels on the screenshot above1. Top left is where you usually see your scripts, R markdown documents or any other opened files. Sometimes you will have a tab with the view of a data frame. Most of the time, this is where you will be working – mostly typing your code in a script or R markdown file. Chances are that you don’t see this panel yet, as you have not opened any files – you will see it in a moment.
1 You can customize that view, of course. You can change colors, position, elements shown and much, much more. Take a look in Tools -> Global Options.
2 The “commands” that you execute in R are properly called “expressions”.
On the top right panel you have several tabs. You will soon be using the “Environment” tab, which shows you the variables that you have created and their values. The “History” tab shows you the history of commands2 that you have typed in the console. There are other tabs here that might come in handy at some point in your work with R – one that you might want to try to supplement this course is the “Tutorial”, which walks you through basics of R (a bit like this book).
The bottom left panel is where the actual R is, or, more precisely, the R console3. The R console is where you can directly access R. If you were to start R standalone (without R studio), the console is the only thing that you would see. You will start typing in the console in a moment, but soon you will learn to indirectly access the console by typing your code in the script and executing it. Note that at the top of the console you can see the path to your project directory. We will discuss paths and directories in a moment.
3 Pronounced /ˈkɒn.səʊl/
Finally, on the bottom right you have, again, several tabs. “Files” is a file browser. You can navigate your file system here, click to load files in R or preview them, and more. In another tab, you will see the plots once you generate them.
Exercise 1.2 (Help) Click on the “Help” tab in the right bottom panel. Type “t.test” in the search bar (mind the dot!) and press Enter. What do you see? Go through the document and notice the structure of the manual page. The majority of the manual pages will have precisely the same structure, as it is a part of the R documentation system.
You can use R as a very powerful calculator. For example, do you want to know what \(\sin(\pi/2)\) is? Just type sin(pi/2) in the console and press Enter. Addition and subtraction work, as expected, with + and -. To multiply two numbers, type 2*3; to divide, type 2/3. You can get exponents (powers, eg. \(2^3\)) by typing 2^3. If the ^ symbol (called “caret”) is not available on your keyboard, you can use ** instead. Parentheses () are used to group expressions, just like in mathematics. To logarithmize, you can use log(), log2() and log10() functions. For example, to calculate \(\log_{10}(100)\), type log10(100). Can you guess how to calculate \(\sqrt{2}\)? Yes, you are right: sqrt(2). Or 2^(1/2), that will also do. Finally, the exp() function calculates the exponential function \(e^x\).
sin()log(), log2() and log10()sqrt()exp()On the left side of the RStudio window you have (by default) two panels: the lower one is called “Console”. When you create a new script file, as you have done it a moment ago, it appears above.
You can type your commands (properly called “expressions”) directly into the console, but it is generally not a good idea. Why? The truthful answer is: because it is messy and sooner or later you will regret it. You can save the history of what you type in the console, but it is easier (and cleaner) to save your program in a script file.
When you open or create an R script file and type something into it, you can send it to console and execute it. To do that, you have two options. First, you can use the “Run” button in the script panel:
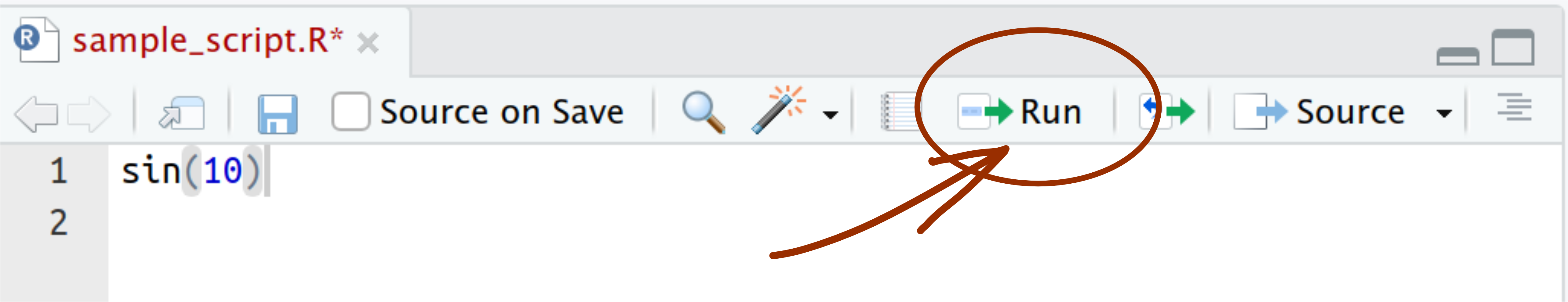
However, this is one of the most common operations, so it is much more efficient to use a keyboard shortcut: Ctrl+Enter (or Cmd+Enter on Mac). This will send the current line of code to the console, where it will be executed, and the cursor in the script will move to the next line of code. You can also select a fragment of the code before you press Ctrl+Enter, and then the whole selected fragment will be sent to the console.
There are many keyboard shortcuts in RStudio. You can see them all in the “Help” menu, under “Keyboard Shortcuts Help”. You can also customize them in the “Tools” -> “Modify Keyboard Shortcuts” menu.
What if we want to store the result of a calculation for later use? We can do this by assigning the result to a variable. In R, you assign a value to a variable using the <- operator:
<- assignment operatorIf you want to see the value of a variable, just type its name in the console and press Enter, or use print() function:
print()Many other languages use = as an assignment operator. In R, you can use = as well, but do yourself a favour and don’t. Use <- instead. Why? Your code will be more readable and you will avoid many common mistakes.
Variables are like boxes in which you can store values. However, unlike boxes, when you assign one variable to another, the first variable keeps its content:
We now come to a very important point which we will revisit often, as it is one of the most common beginner (and not only beginner) mistakes. When you forget to assign the value to a variable, R will print it to the console, but the variable will not be modified:
In the code above, the value of x is not changed by the sin() function. To store the value of a function, you need to assign it to a variable:
Please spend some time on this, as it is a very common source of errors.
Exercise 1.5 Without actually running the code, guess what will be the value of x if you execute the following code:
Just run the code ;-) Which lines contain the assignment operator?
As a rule of thumb4, if the expression you type in your script does not contain the <- operator, it will not modify any variables.
4 There are exceptions to this rule, but they are relatively rare and I will not discuss them here.
Exercise 1.6 Create a variable using x <- 42. Take a look at the Environment pane in RStudio (top left part of the window). Do you notice anything?
A new entry appeared in the “Environment pane”. It shows that there is a new variable present in your environment.
Variables can store not only numbers, but also text. Text in R is called a character string. To create a character string, you need to enclose the text in quotes (both single and double quotes are allowed, but try to be consistent and use only one type). For example:
Character variables cannot be used with algebraic computations, the following code will throw an error:
However, if you want to “add” two character strings (that is, concatenate them), you can use the paste() function:
paste()Quite often, you don’t want to have a space between the two strings. This is such a common operation that R has a shortcut for it:
paste0()There are other types of data types in R. Later on, we will briefly touch on factors, which look like character strings but behave like numbers. Another important data type is a logical type, which can have only two values: TRUE and FALSE. We will talk about logical types in more detail tomorrow. And under the hood, numeric vectors can be either integers (numbers like 1, 2, …) or floating point numbers (numbers like 1.1, 2.2 or \(\pi\)).
Exercise 1.7 (Variables) Create the following variables in your script:
name with the value of your first namecity with the value of the city where you liveage with the value of your agegreeting with the value “Hello,”greeting and name and store the result in a new variable hellothereWhen you work with R, you create variables, functions and other objects. They appear in the “Environment” tab in RStudio and constitute what is known as a workspace.
When you exit R, for example when closing RStudio or switching to another project, R / RStudio will ask you whether you wish to save the current workspace and / or history. This can create two files in your project directory: .RData and .Rhistory (or _RData and _Rhistory on Windows). The .RData file contains the workspace, that is, all the objects (variables, functions, etc.) that you have created. The .Rhistory is a text file (you can open it in any text editor) that contains the history of commands that you have typed in the console.
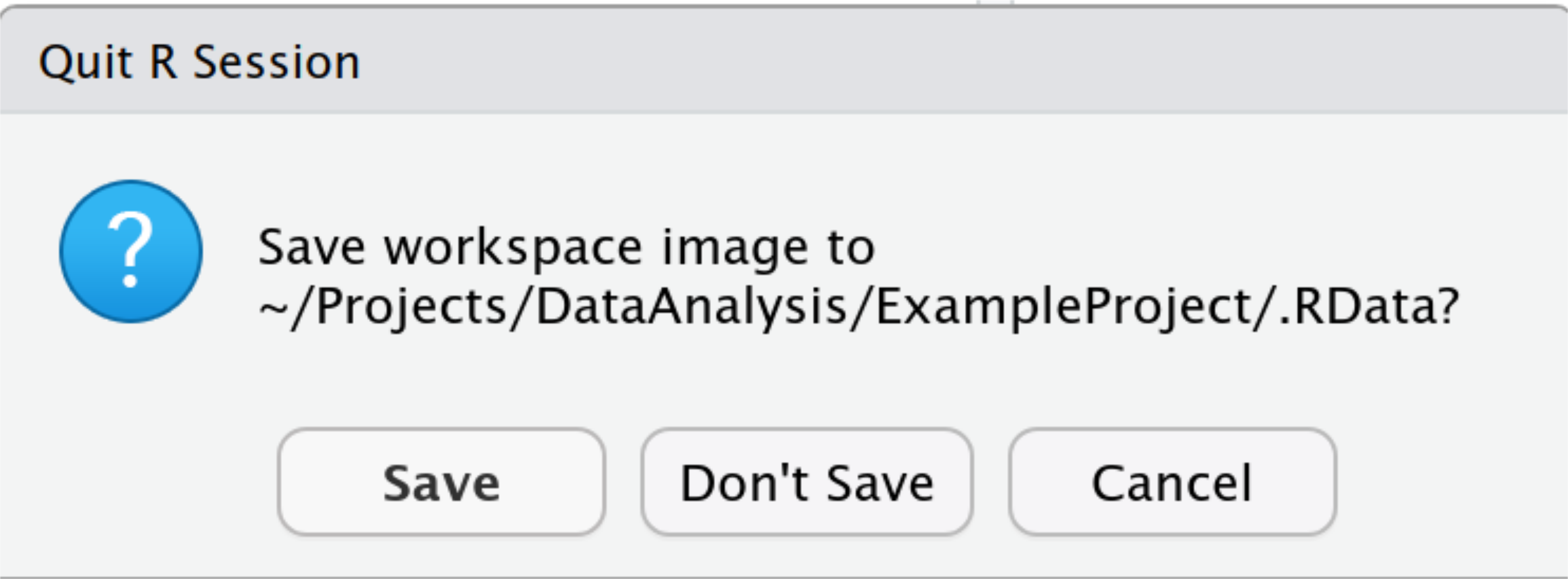
That all sounds like a good and useful thing, right? By saving the workspace, you do not have to repeat all your calculations! And by saving the history, you don’t even need to type your code in a script, since R saves everything automatically, right?
Well, not so fast. Relying on that can get you into trouble.
Workspace. Saving a workspace can be useful, but it can also get you in a mess. When working with R interactively, one tends to create a lot of objects just to try out various things. Not all of them will go into your final version of the script, but the mere fact that they exist in your environment can be a problem: they can interfere with your code, take up memory and even can hide certain bugs.
Consider this example: trying out various things, you create a variable called foo. Later on, in a much later version of the script, you no longer create foo, you now have a “proper” name like mouse_transcripts. However, one of your script function still uses foo instead of mouse_transcripts. You run the script and it works! But it works only as long as you have the foo variable in your workspace. If you were to run the script on a different computer, or give it to your colleague, it would stop working.
History. Beware! By default, R saves only the last 1000 lines of your history.
Do not rely on the history and workspace to save your work. Always save your scripts, and if you want to save the data, save it in a separate file.
Variables can (and do) store a lot more than single values. One of the most basic and important data types in R is a vector. A vector is simply a sequence of values – just like in maths. And you know what? You have already created vectors in R. In mathematics, any scalar value can be treated as a one-dimensional vector and it is exactly like that in R: any single value is a 1-element vector, including all the variables that you have created in the previous exercise.
To create a vector with more than one value, you can use the c() function (“c” stands for “combine”). For integer numbers, you can use the : operator to create a sequence of numbers. For example:
c()Creating a sequence of numbers with :It is also possible to combine two vectors longer than 1 into one:
[1] 1 2 3 4 5Exercise 1.9 (Vectors) Create a vector that combines the numeric value 1 and the string "one". What happens? Can you venture a guess?
You can access individual elements of a vector using the [ ] operator:
[ ]But hey, I told you that every value is a vector in R, right? And that includes the indices 1 and 2 that you have just used. So, what would happen if we used more than two values as an index? Try it:
[1] 10 42 33[1] "Hoppegarten" "Weiner" "January" [1] 10 25 33As you can see, not only can you use a vector as an index, but you can also use a variable as an index.
It is tempting to select, say, first and the third element of a vector numbers by writing numbers[1, 3]. This will not work! As you will see tomorrow, this way of writing is for two-dimensional objects. You must use a vector as an index: numbers[c(1, 3)].
In many (most?) programming languages, the first element of a vector is accessed using the index 0. For example in Python, to access the first element of an array, you need to type array[0]. This has something to do with how computers work. In R, the first element is always 1 – R was designed by statisticians, and in mathematics we always start counting from 1. For some reason, this seems to make some computer scientists angry.
Accessing elements of a vector using indices is all well and good, but sometimes it can be very inconvenient, especially if the vectors are very long. Or maybe you do not remember the order in which you have stored the elements of the vector – was the last name first, or second element of the person vector?
Vectors allow you to name their elements. We can either define the names at the very beginning, when we create the vector, or we can add them later using the names() function. Here is how you can do it:
Once you have named the elements of a vector, you can access them using their names:
Or, we can change the names with the names() function:
names()OK, one more thing about vectors. Above we have selected elements from a vector. It turns out, we can do more with that selections then just print it to a console:
[1] 100 42 33 14 500Here is what happened: we assigned new values to the first and the fifth element of the vector numbers. This is a very powerful feature of R and you will be using it a lot.
Exercise 1.10 (Accessing and modifying vectors)
primes.primes[11]?primes[11] <- 31?primes[15] <- 47?primes[-1]?# note that 1 is not a prime number!
primes <- c(2, 3, 5, 7, 11, 13, 17, 19, 23, 29)
primes[c(3, 5, 7)][1] 5 11 17[1] NA [1] 2 3 5 7 11 13 17 19 23 29 31# adds a new element to the vector
# at position 15, fills the gap with
# NAs
primes[15] <- 47
primes [1] 2 3 5 7 11 13 17 19 23 29 31 NA NA NA 47 [1] 3 5 7 11 13 17 19 23 29 31 NA NA NA 47Vectors are very useful – but wait, there is more. What happens if we add a value to a vector? Try it:
As you can see, R has added the value 10 to every single element of the vector numbers. The same thing happens with other operators, like -, * and /. Try it yourself.
This is called vectorization and it is one of the most powerful features of R compared to other languages. It will allow you to write very concise and, at the same time, readable code.
The vectorization works not only with operators like +, -, * and /, but with many functions. For example, it works with most of the mathematical functions like sin() or log(). Try it:
[1] 1.000000 1.623249 1.518514 1.146128 1.397940[1] -0.5440211 -0.9165215 0.9999119 0.9906074 -0.1323518However, there is a catch. What happens if you try to add two vectors when both of them with more than one element? First, let us try to add two vectors of the same length:
As you can see, R has added the first element of the first vector to the first element of the second vector, the second element of the first vector to the second element of the second vector, and so on. Makes sense, right? Same would happen if we were to subtract, multiply or divide the vectors (or use logical operations, which you will learn on Day 3).
Imagine the two vectors one beneath the other:
numbers1: c(1, 2, 3)
+ + +
numbers2: c(4, 5, 6)
↓ ↓ ↓
result: c(5, 7, 9)R is simply adding up corresponding elements. This does not look like much now, but trust me, it will be extremely useful in the future.
However, if the vectors have different lengths, it is a different story altogher. Take a look:
Warning in numbers1 + numbers2: longer object length is not a multiple of
shorter object length[1] 5 7 7Ooops, what exactly happened here? First thing to note is that there was no error. There was a warning, but still our code executed and produced a result. But what is that result? For the first element of the result, it is clear enough: 1 + 4 = 5. Same for the second, 2 + 5 = 7. But what about the third? It seems that R added 3 + 4 = 7. But why?
R noticed that it is missing an element to be added to the third element of the vector numbers1. So, it did what is called recycling. It “rewound” the vector numbers2 to the beginning and added the first element of numbers2 to the third element of numbers1. However, since after the rewinding and adding one element of vector numbers2 was left (because numbers1 did not have any more elements), R issued a warning.
numbers1: c(1, 2, 3)
+ + +
numbers2: c(4, 5) c(4, 5)
↓ ↓ ↓
result: c(5, 7, 7)
If the length of the first vector was a multiple of the length of the second vector, R would not have complained:
See? No warning. R was recycling the second vector over and over again. Recycling is a dangerous business: if you are not careful, you can get results which you have not expected.
numbers1: c(1, 2, 3, 4, 5, 6)
+ + + + + +
numbers2: c(7, 8) c(7, 8) c(7, 8)
↓ ↓ ↓ ↓ ↓ ↓
result: c(8, 10, 10, 12, 12, 14)
Take it slow. This is advanced stuff, but I had to warn you already at this stage – this is one of the common sources of errors in R. Watch out for this “longer object length is not a multiple of shorter object length” warning.
Here is my advice to you when using vectorization: either use a vector and a single element vector, or two vectors of the same length. And in the cases where, for some reason, you need to recycle, make sure that you know what you are doing. For example, check the length of both vectors.
With vectors that have only a couple of numbers it is quite easy to see what is happening, but what if you have thousands of variables? In other words, how to check the lenght of a vector? You can use the length():
length()Exercise 1.11 (Vectorisation)
lengths, contains the lengths of the sides of three rectangles, and the other, widths, contains their widths. Create a vector containing the areas of these rectangles.[1] 11 12 13 14 15[1] -9 -8 -7 -6 -5[1] 10 20 30 40 50[1] 0.1 0.2 0.3 0.4 0.5[1] 0.7853982 19.6349541 132.7322896[1] 3[1] 4 10 18R has three types of information to pass to you: messages, warnings and errors. Messages are just that – messages. Warnings are messages that tell you that something might be wrong and you should pay attention, but R will nonetheless do what you asked it to do. Errors stop the execution of your code, but warnings do not.
You should pay attention to warnings, but you do not have always to do something about them – some you can safely ignore. Errors, on the other hand, you should always fix.
NAOne more thing: there are a couple of special values in R that you should know about. One of the most prominent, useful and frequently encountered is the NA value, which stands for “Not Available”. You will see it frequently when you work with data. Actually, you have already seen it when you tried to access an element of a vector that does not exist.
NAIt is possible to apply mathematical operations to NA values, but the result is inadvertently NA:
This also goes for some functions, which, quite often, have a special argument to omit the NA values. For example, the mean() function calculates the mean of a vector:
mean()There is a whole bunch of functions that you can use to work with vectors, and here are some of them – with mostly self-explanatory names: sum(), min(), max(), range(), sd(), var(), median(), quantile(). Look them up in the help system by typing, for example, ?sum in the console, and try them out to see how they work.
sum(), min(), max() etc.The NA values very frequently pop up when you try to convert a character vector holding what looks like numbers into a numeric vector. We will see many such examples in the days to come; the conversion is often done using the as.numeric() function. For example, it is quite common that values typed in a spreadsheet contain comments or values which look like this: > 50 (and means: measurement out of range).
as.numeric()imported_data <- c("10", "20", "30", "> 50", "40", "N.A.", "60 (unsure)")
# the following will generate a warning
imported_data <- as.numeric(imported_data)Warning: NAs introduced by coercion[1] 10 20 30 NA 40 NA NAAs you can see, R conveniently warns you that some elements of the vector were changed to NA. Look out for that warning!
There are a few other special values in R that behave similarly to NA. Inf stands for infinity, you will get it when you divide a positive number by zero: 1/0. -Inf is the negative infinity (when you divide a negative number by 0), and NaN stands for “Not a Number” – this is what you get when you try to subtracting Inf - Inf or dividing 0/0. They have also other uses – for example, if a function wants to know how many rows of output you would like to see, and your answer is “all of them”, you can use Inf as the number of rows.
Inf, -Inf and NaNThere is an old puzzle that goes: “On a lake, there is a water lily. Each day the lily doubles in size. After 30 days, the lily covers the entire lake. On which day was the lily covering half of the lake?”.
In the following section we will model the behavior of the lily using R. Let us start with some assumptions.
Exercise 1.12 (Modelling water lilies) Take a piece of paper and a pen. Your task is to come up with a formula to describe the area of the lily on the \(n\)-th day. Write down the formula.
Hint: if you are stuck, try to calculate the area of the lily on the first few days.
There is an important point that we wish to demonstrate here. Quite often it pays off to close your laptop and think for a moment what it is it that you want to do, rather than start coding right away. Pen and paper are helpful (we will be making this point again when it comes to visualizations). If you do not have a clear idea of what you want to do, you can get stuck thinking about what you already know how to do.
The formula for calculating the area of the lily on the \(n\)-th day is \(0.01 \times 2^{n-1}\). You can come up with that result quite easily if you consider the first few days. On day 1, the area is \(1\% = 0.01\). On day 2, is twice that, that is, \(0.01 \times 2 = 0.01 \times 2^1 = 0.02\). On day 3, it is twice the area from the previous day: \(0.02 \times 2 = 0.04 = 0.01 \times 2 \times 2 = 0.01 \times 2^2\). And again, on day 4, it is \(0.01 \times 2^3\). And so on5. We can show it in a table:
5 If started counting from 0 – that is, if we designated the first day as Day 0 – the formula would be \(0.01 \times 2^n\).
| Day | Area | Calculation | Formula |
|---|---|---|---|
| 1 | 0.01 | \(0.01\) | \(0.01 \times 2^0\) |
| 2 | 0.02 | \(0.01 \times 2\) | \(0.01 \times 2^1\) |
| 3 | 0.04 | \(0.01 \times 2 \times 2\) | \(0.01 \times 2^2\) |
| 4 | 0.08 | \(0.01 \times 2 \times 2 \times 2\) | \(0.01 \times 2^3\) |
| 5 | 0.16 | \(0.01 \times 2 \times 2 \times 2 \times 2\) | \(0.01 \times 2^4\) |
Once we have the formula, it is very easy to calculate the area covered by water lillies on the first 10 days. We will use vectorization to do this:
This calls out for a plot. We will talk about visualizations more extensively on Day 5, but for now, we will use a very basic and simple function to plot the area of the lily on the first 30 days. The function is called plot() and can be used to plot a graph of two vectors. The first vector is the days, the second vector is the area.
O-K, days and area are clear, but what is this type="b"? This is a so-called named argument6. The plot() function has many arguments, and if you want to use only some of them, you can use their names with an equal sign. You will see that a lot in the days to come. This particular argument, type, tells R what kind of plot to draw. The "b" stands for “both” and tells R to draw both points and lines. If you want only points, you can use "p" (or simple leave the argument out), if you want only lines, you can use "l".
6 Full disclosure: all arguments have names in R and can be named explicitely. However, some of the arguments have a default value, so we do not have to specify them unless we need them. The type argument is one of them. Others must always be specified.
Note another thing on this plot: after day 7, the area is greater than 1. But 1 means 100%, so after day 7, the lily covers more than the entire lake. Obviously, this is not possible – and it shows a limitation of our model. We can show it by adding a horizontal line to the plot:
Exercise 1.13 (Plotting water lilies)
plot() function, but add, as a parameter, col="blue". What happens?pch=19. What happens?xlab to label the x-axis with “Day”. Use the argument ylab to label the y-axis with “Area”. Use the argument main to give the plot a title.ylim=c(0, 1) to change the range of the y-axis. How would you change the limit of the x axis?area_slow) which will be calculated with the new formula.lines(). What does the lines() function do? (Hint: type ?lines in the console).(Solution)
R is a so-called functional language. This is different from many other languages (including Python). It has some interesting implications which we will partially explore over the next few days. For now, however, we will be content with one important statement: in R, most of the stuff you do, you do using functions. A function takes zero or more arguments and returns exactly one argument:
During this course, we will not really discuss or require creating your own functions. However, I would nonetheless like to show you how it is done. There are two reasons for that. Firstly, it is really, really easy. Secondly, it will help you understand how functions work in R, and that will help you understand how to use functions that others have created.
In the water lilies example we have used a formula to calculate the area of the lily on the \(n\)-th day. The formula includes three parameters: the initial fraction of the area covered by the lily on day 1, the day number and the factor by which the area is increased each day. We will now create a function that takes two parameters: the day (or days) and the factor, and return the area of the lily on that day. Here is how you can do it using function() keyword in R:
function() { ... }As you can see, the function is created using the function keyword. In parentheses (( and )), you specify the arguments that the function takes, separated by commas. Then comes the body of the function, enclosed in the curly braces ({ and }). On the last line of the function code, the return() function is used to return the value of the area_lily function.
Once you have run the code above, you can use it to calculate the area of the lily on the first 10 days like this:
One interesting and important fact about the defining the functions is that you use the assignment operator <- to assign the function to a variable. In other words, area_lily is, in fact, a variable! A variable which holds not a value or character string, but computer code that can be used to do stuff. You can copy it to another variable and it will behave exactly as the original function:
Exercise 1.14 (Creating your own function) Modify the area_lily function so that it takes three arguments: the day, the initial fraction and the factor. Use the new function to calculate the area of the lily on the first 10 days with the initial fraction of 0.001 and the factor of 1.5. What is the area on the 10th day?
It is now time to conclude today’s lesson with a bit of philosophy. When you write an R script, the first goal you have in mind is to analyse your data – in other words, by means of what you write you are trying to make the computer do something for you. That is correct and fine, but there is an important aspect of programming that is often overlooked.
When you write a program, you are writing it not only for the computer, but also for other people. These other people may include your colleagues, readers of your scientific articles, your students, and, last but not least, a future version of yourself. All these people need more then just a piece of code that works. You will quickly find it out yourself when you open a script or a project that you have not been working on for a few months – trust me on this, you will not know what it does, how it does and sometimes even whether you have written it yourself or copied from somewhere.
You might be thinking that you are never willing to show your code to another person. You are wrong, and not only because it is useful to you for another person to review your code. Firstly, you will want to share your code because as a scientist you will want to share your results, and results are nothing if the methods to obtain them are unknown. And secondly, you will need to share your code because you will be asked to do so by your colleagues (yes, I was as surprised as you will be when I was asked to share my code for the first time). And thirdly, your code is part of your methods and you will have to share it when you publish your results7.
7 Top journals already require that you share your code when you publish your results. This will become more and more common in the future.
For communication with another human being to be efficient, you need to make it as clear as possible. There are several ways how to make your code more readable and understandable. Here are some of them.
Comments. Comments are lines in your code that are not executed. In R, they start with a # sign. Comments help to explain what exactly are you trying to achieve with your code. The old saying goes: “Code tells you how, comments tell you why”. You can hardly overdo with comments, but you can easily underdo.
#Naming. The names of your variables, functions and files should be meaningful. If you have a variable that stores the number of days, call it days, not x. If you have a function that calculates the area of a circle, call it calculate_circle_area(), not f(). If you have a comma separated values (CSV) file that contains the CRP values, call it crp_values.csv, not data.txt. This is sometimes much more difficult than it looks, but it is very important. Also, that does not mean that you can’t use short names – but use it only for “throwaway” variables that are used only once or twice, or for example code.
Formatting. Your code should be formatted in a consistent way. For example, you should always put a space around your operators, like x <- 2 (and not x<-2), you should always put a space after a comma, like c(1, 2) and not c(1,2) (and also not c( 1 , 2 )). Lines should not be too long – 80 characters at most is a good rule of thumb. If a line is too long, you can split it into several lines – unlike Python, R will not mind. See here for a more detailed guide on how to format your code.
The following fragment of code shows how you should not format your code:
The code is correct, but it is hard to read. What does it do, quickly? If you carefuly examine it, you will see that it calculates the standard deviation of the vector b, following the formula
\[SD = \sqrt{\frac{\sum_{i=1}^{n} (x_i - \bar{x})^2}{n-1}}\]
where \(n\) is the length of the vector b, \(x_i\) is the \(i\)-th element of the vector b and \(\bar{x}\) is the mean of the vector b. However, there are several issues.
Firstly, there are no comments in the code which would give a hint of what it does. Secondly, \(n - 1\) (the variable a) is hard encoded - if you modify the vector b by adding one number, the code will execute, but the result will be incorrect.
Thirdly, line 4 combines several operations making it very hard to read. It should be split for clarity. The following code is much more readable:
# ---------------------------------------------------------
# Calculating the standard deviation of a sample
# ---------------------------------------------------------
# example values for five samples
samples <- c(1, 10, 20, 21, 5)
samples_n <- length(samples)
# calculate standard deviation of samples manually
samples_mean <- mean(samples)
samples_devs <- samples - samples_mean
# samples variance
samples_var <- sum(samples_devs^2) /
(samples_n - 1)
samples_sd <- sqrt(samples_var)This makes it absolutely clear what you are trying to do, and, in addition, calculates the mean, the deviations, sample length and sample variance – all of which might come in handy later on. Also note of the use of # ----… comments. Programmers often use these to highlight the beginning of a new section of code. This is not necessary, but adds to readability. Lines 14 and 15 show how you can split a line of code in a way that is both readable and clear.
Of course, the example is a bit silly – R has a lot of built-in statistical functions, and standard deviation naturally is one of them. You can calculate the standard deviation of a vector b using the sd() function:
sd()Nonetheless, the principle stands.
OK, so you need to use explicit variables. Isn’t that a bit cumbersome? A lot of typing? An opportunity to make typos?
Not really, thanks to a bit of magic called “tab completion”. Say, you have defined a variable samples_sd:
Now you would like to display it. You start typing sam and press the tabulator (Tab) key or simply wait for an eyeblink, and then R will show you a list of all variables and functions that start with sam. You can then use the arrow keys to select the one you want, and press Enter or Tab to insert it into your code.
This is an extremely useful feature. You will use it a lot and there is way more to it. For example, if you start typed a function name with the opening parentheses, like sd(, then pressing “Tab” will show you all available arguments of the function, and you can scroll through them to see what they do. Go on, try it!
Things that you learned today:
# signpaste() and paste0()c() and :[ ]sqrt(), log(), log2(), log10(), sin(), cos(), tan()sum(), mean(), min(), max(), range(), sd(), var(), median(), quantile()NA, Inf, -Inf, NaNTRUE and FALSEplot()XXX
1.4.3 Comments
If you start your line with
#(called “hash” or “pound” sign), the rest of the line will be ignored by R. This is called a comment and I will spend some time later on convincing you that you should use a lot of comments in your code.Comments are also a great way to temporarily disable a line of code - we call it “commenting out”. This is for the cases when you want to try out something, but you do not want to delete a line of code that may be still useful later on.
Exercise 1.4 Repeat Exercise 1.3, but now type the expressions into the script which you have created in Exercise 1.1. Before each expression, insert a comment line stating what it does, for example
# calculate sinus of pi. Run the script by pressingCtrl+Enterafter each line.Script files are also text documents. You can open them in any text editor, for example Notepad or even Word (but don’t do that). In RStudio, you see the script file in many colors: for example, comments can be green, strings (text in quotes) can be red, and so on. This is called syntax highlighting and is done by RStudio to make your code more readable. You will not see the colors when you open your R script in Notepad.
From now on, you should only type your code in script files.